MiRabel is a webtool dedicated to the efficient prediction of miRNA targets.
miRabel can predict mRNAs that are targeted by several miRNAs given as input.
As an option, the experimental expression level of each miRNAs is taken in
account to evaluate their potential cumulative effect on the mRNA targets.
In addition, results are linked to metabolic and cellular pathways retrieved
from KEGG which allows a more comprehensive analysis of the miRNAs.
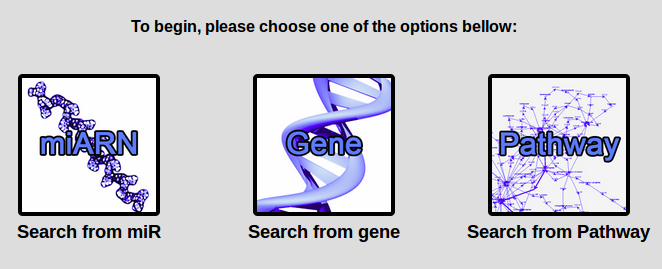
There are two ways of getting to the predictions pages.
First of all, you can use one of the 3 links included in the submenu of
the "get started" menu bar.
Otherwise, you can click on any of the 3 links available on the home page.
They correspond to 3 different search types:
- The "by miR" search
- The "by gene" search
- The "by pathway" search
For information on each search type, see their respective section.
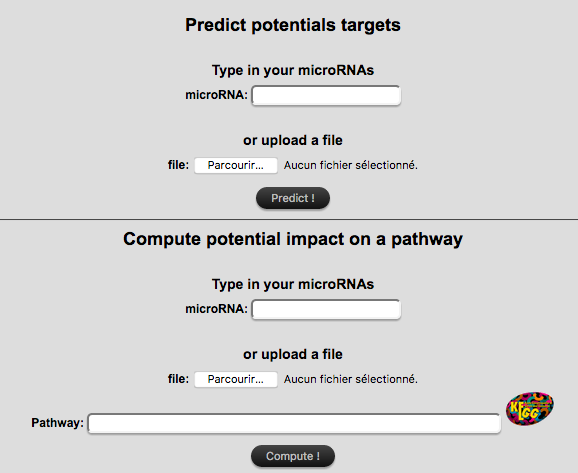
The "by miR" search requires one or more miR(s) as entry (and optionally their associated expression
factor) and returns a list of potentially targeted genes with their associated scores.
MiRs can be directly typed in the text field or uploaded
(
TSV
format, composed of one column with miR names and optionally a column with expression factors).
It is also possible to restrict predictions to a specific pathway of interest.
Click "Predict !" or "Compute !" to launch the search.
- IMPORTANT:
If you use both the text fields and the file input field at the same time, only the file data will be treated.
Information in text field will be lost.
Moreover, providing expression factor for some but not all micro-RNAs may result
in wrong or unexpected results.
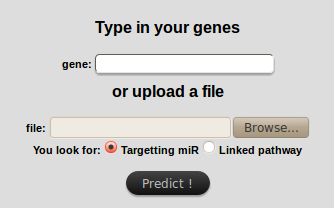
The "by gene" search requires one or several gene(s) in entry.
Genes can be directly typed in the text field or uploaded
(
TSV format,
composed of one column with gene names).
You then need to choose whether you want to get the potentially
targeted miRs or the list of pathways the genes you entered are involved with.
Hit "Predict !" to launch the search.
- IMPORTANT:
If you use both the text fields and the file input field at the same time,
only the file data will be treated.
Information in text field will be lost.
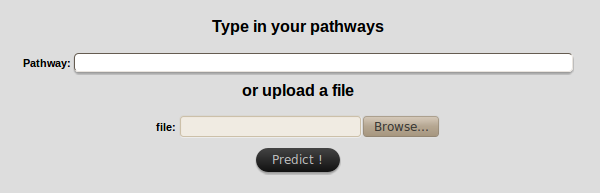 The "by pathway" search requires one or more pathway(s) (with a maximum of 10)
to get the list of genes that they contain.
Pathways can be directly typed in the text field or uploaded
(TSV format,
composed of one column with pathway names).
The "by pathway" search requires one or more pathway(s) (with a maximum of 10)
to get the list of genes that they contain.
Pathways can be directly typed in the text field or uploaded
(TSV format,
composed of one column with pathway names).
- IMPORTANT:
If you use both the text fields and the file input field at the same time,
only the file data will be treated.
Information in text field will be lost.
Moreover, if using the file input field, make sure that the pathway names you entered
match exactly those included in KEGG database!
The converter is accessible from the "Tools" menu bar item.
Type in your miR name(s) of interest and hit "Convert !".
You will get the most recent nomenclature for the given miRs as well as their associated miRbase version.
The search formulars are not extending and / or data is not / badly display.
This application uses javascript.
Please verify that you have enabled java in your browser.
Often, you can find this option in "Edit" or "Preferences" on your browser.
When exporting result, only displayed data is saved.
If you wish to export the entire dataset, choose to display all rows in the "display length selector"
before exporting.
When doing a search, no results are found.
Please check the spelling of your entries.
It is also possible that no prediction exist in the database for your entry.





 The "by pathway" search requires one or more pathway(s) (with a maximum of 10)
to get the list of genes that they contain.
Pathways can be directly typed in the text field or uploaded
(
The "by pathway" search requires one or more pathway(s) (with a maximum of 10)
to get the list of genes that they contain.
Pathways can be directly typed in the text field or uploaded
(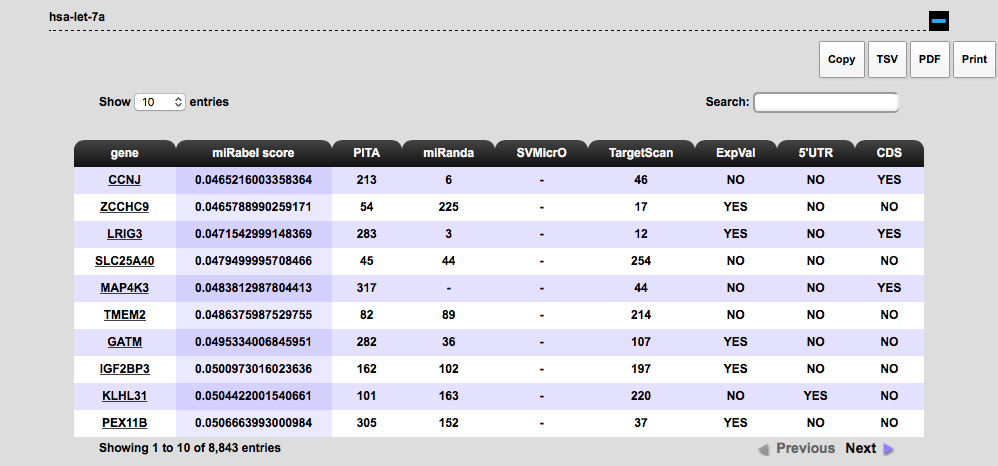
 You can export your data (top right corner) in 4 different formats:
You can export your data (top right corner) in 4 different formats: